Speed Genomic Analysis by Boosting Accelerator Utilization and Lowering Costs
What Could 32 GPUs in a Single Node Do For You?
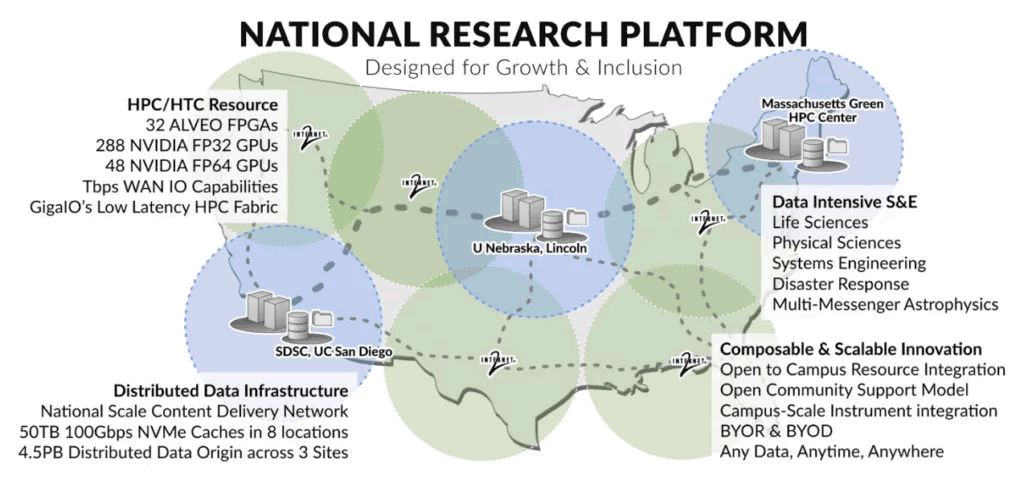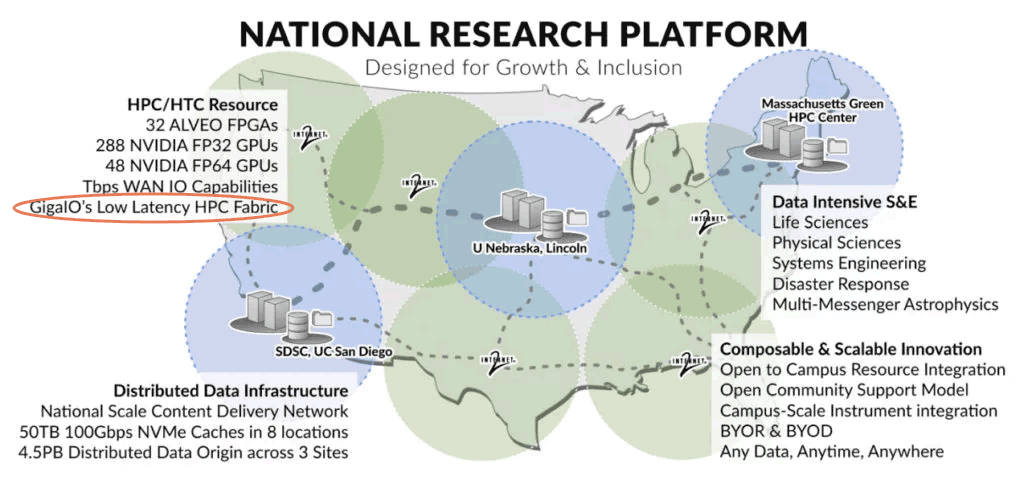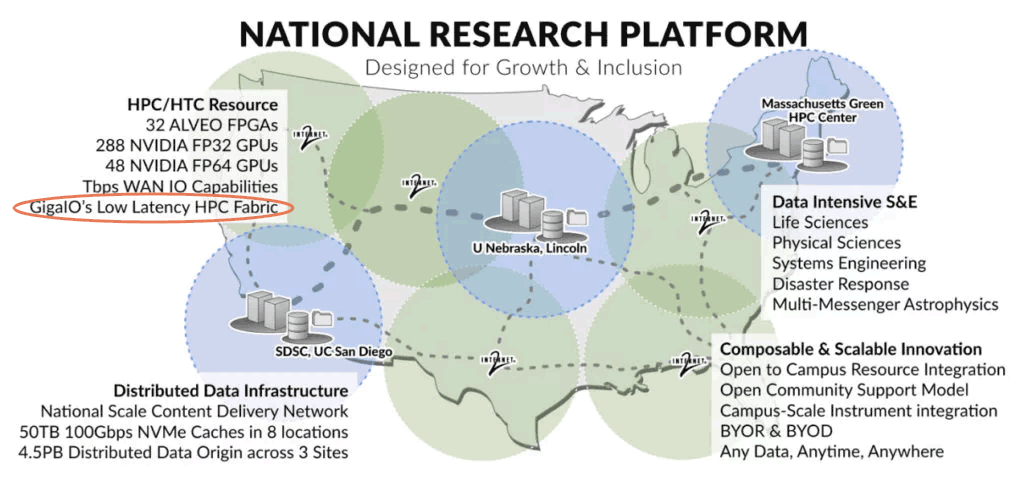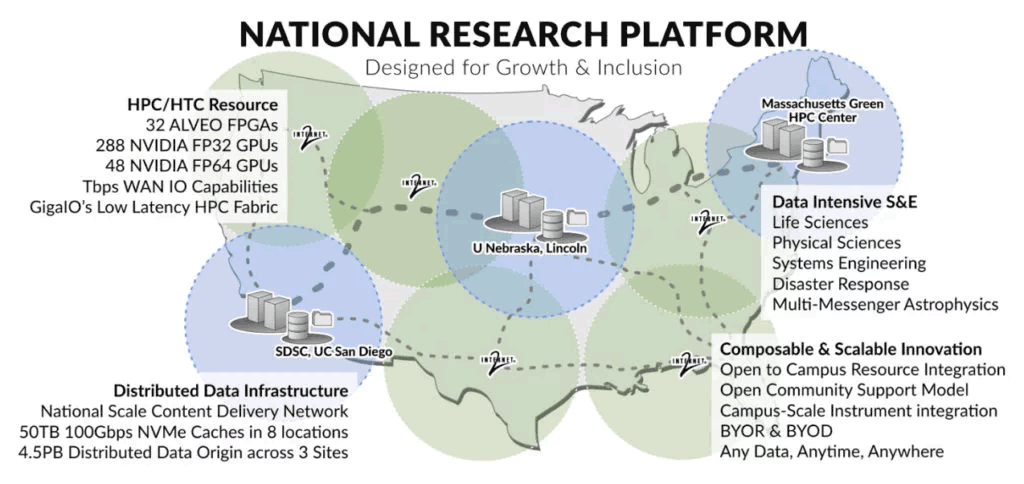
SDSC Genomics Processing & Analysis: Prototype National Research Platform

UC San Diego’s Tianqi Zhang and Tajana Rosing, one of the co-principal investigators of the Prototype National Research Platform (PNRP), developed applications that run on FPGA accelerators for basic genomics processing components, like sequence trimming and alignment, and integrated them with the pipelines for COVID-19 phylogenetic inference, microbial metagenome analysis and cancer variant detection.
“It’s pretty easy to migrate the previous programs to the new U55C cluster [PNRP]. The development platform is also similar to the local environment, with only a few board configurations needing administrator intervention. We are currently scaling up and optimizing the accelerators on the multi-FPGA nodes. If successful, it will provide O(10x) speedup and O(100x) power savings compared to CPU,” said Zhang.
“Our research requires that we aggregate disparate computational elements into highly usable and reconfigurable systems. FabreX makes it possible to dynamically bring these elements together in a very low-latency, high-performance interconnect while allowing for distinct, non-interfering workflows to co exist on the same infrastructure.”
— Dr. Frank Würthwein, Director